data base part of the GMD
GMD started as a collection of annotated and non-annotated, but repeatedly observed mass spectra from biological samples and was extended to contain, in addition, retention time behavior (Wagner et al 2003). Subsequently, the concept of mass spectral tags (MSTs) was developed (Kopka 2006). This concept became necessary because commercially available mass spectral libraries, such as the national institute of standards and technology (NIST) standard reference database, NIST05 (http://www.nist.gov/srd/nist1a.htm) with the NIST MS search software version 2.0 (http://chemdata.nist.gov/mass-spc/Srch_v1.7/index.html) and Wiley mass spectral library 2005 (http://eu.wiley.com/WileyCDA/WileyTitle/productCd-0471755958.html), only contained a small fraction of those compounds that were frequently observed in profiling experiments of biological samples. The MST concept allows handling and referencing of yet unidentified metabolic components from GC-MS profiling experiments. Also, MST collections allow later identification using pure authentic reference substances. Today, multiple laboratories have added to and complemented the initial library content (Schauer et al. 2005). The current focus is the analysis of approximately 1,000 commercially available reference substances representing metabolites for the purpose of enhanced MST and metabolite identification. In addition, integration of metabolite profiling data with 7 external databases such as KEGG or visualization tools, e.g. MapMan (Thimm et al. 2004), is in preparation.
schema
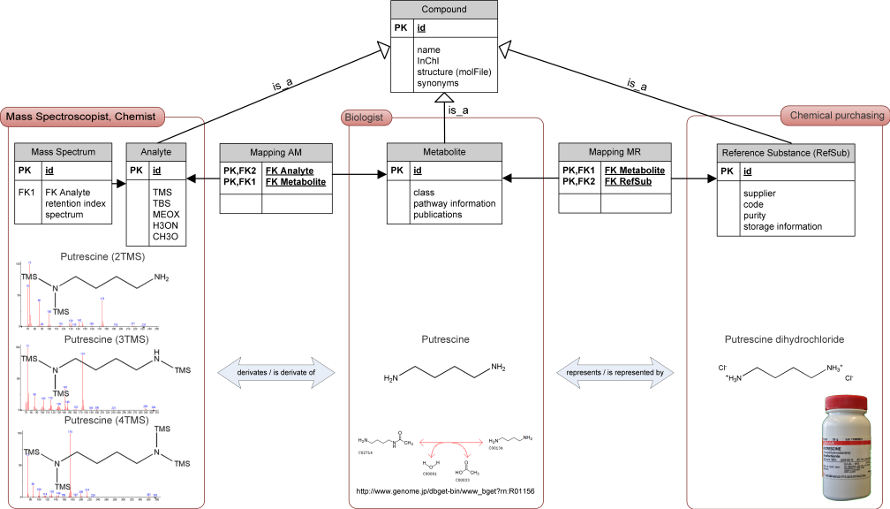
Metabolite profiling experiment can be considered multi-disciplinary involving areas of biological, analytical and chemoinformatic expertise. A biologist is typically interested in the actual metabolite, which is the biologically active substance under investigation and which also relates to metabolic pathways or signalling phenomena. However, in order to perform GC-MS profiling analyses, polar metabolite extracts have to be chemically converted, i.e. derivatized into less polar and volatile compounds, so called analytes. For this purpose N-methyl-N-trimethylsilyltrifluoracetamid (MSTFA) or other suitable derivatization reagents, such as N,O-bis(trimethylsilyl)trifluoroacetamide (BSTFA), N-methyl-N- bis(trifluoroacetamide) (MBTFA) or borontrifluoride (BF3) can be used. Frequently, a methoxyamination step is applied prior to the silylation step in order to reduce the number of signals resulting from sugar isomerisations. Subsequently, the resulting analytes are subjected to a gas chromatograph coupled to a mass spectrometer, which records the mass spectrum and the retention time linked to an analyte. Thus, what is measured is a modified form - the analyte - of the actual metabolite. Also, metabolites often cannot be obtained in their respective native biological state. For example reference compounds of organic acids may be only acquirable as salts. Because of such and other examples we introduced the chemical entity of a reference substance. As the analytical grade and purity of a reference substance may vary, we also store the commercial acquisition information, which refers to the supplier, the respective supplier code and lot number. The mapping between analytes and metabolites (A↔M) as well as between metabolites and reference substances (M↔R) are by their nature many-to-many relations. The chemical derivatisation step may produce more than one variant, and more than one reference compound may exist as it is possible to order multiple reference substances of the same metabolite. For example, the metabolite putrescine has a set of different associated analytes, all with full mass spectral and retention descriptors. Putrescine is linked within GMD to three different silylation products, namely putrescine (2TMS), putrescine (3TMS) and putrescine (4TMS), the three typical silylation products of this metabolite. In rare cases, one analyte might have two or even more metabolites associated, which are chemically unstable and convert into the same analyte under the given analysis conditions. The actually measured data, MSTs, are properties associated with analytes. Analytes may be identifiable when their MSTs can be mapped to reference substances. Otherwise, they may carry the status of non-identified. Because of the many-to-many relationships, it is clear that the matching-based annotation step is non-trivial and prone to errors. It was our intention to link within the GMD reference spectra and the retention behaviour of analytes to the respective metabolite and in parallel to the corresponding reference substance, so as to establish a complete chain of evidence for future metabolite profiling experiments. We thereby facilitate the re-annotation of presently unknown analytes and MSTs, track potential errors due to impure reference substances and enable the identification by reference substances which may become available in the future. The replicate mass spectra and RIs are empirically determined using different mass spectral technologies, e.g. time of flight, quadrupole or ion trap based mass detections, and variations of gas chromatographic systems (Strehmel, et al., 2008)
ADO.NET dynamic data tables
current database statistics
(all numbers include stable isotop labeled compounds)Analyte
- Is the derivatised metabolite that is measured by GC-MS.
- 4,663 Analytes
- 3,568 Analytes having valid spectra associated
Metabolite
- Is the compound of interest.
- 2,226 Metabolites
- 2,021 Metabolites having valid spectra associated
Reference Substance
- Is a purchased compound which may not be exactly identical to the metabolite. For example, organic acids may be obtained either as salts or as free acids.
- 3,689 Reference Substances
- (1,705 unique, in house available)
Mass Spectrum
- constituted by masses and intensities.
- GMD Spectrum News Feed
Get involved with latest spectra submitted to the GMD! - 26,590 Spectra in total
- 11,680 Spectra linked to analytes
- 9,156 Spectra linked to analytes and tagged with an RI (retention index)
ADO.NET Data Services Framework aka REST (Representational State Transfer)
- formerly "Project Astoria"
- gmd online and restful


